Examples¶
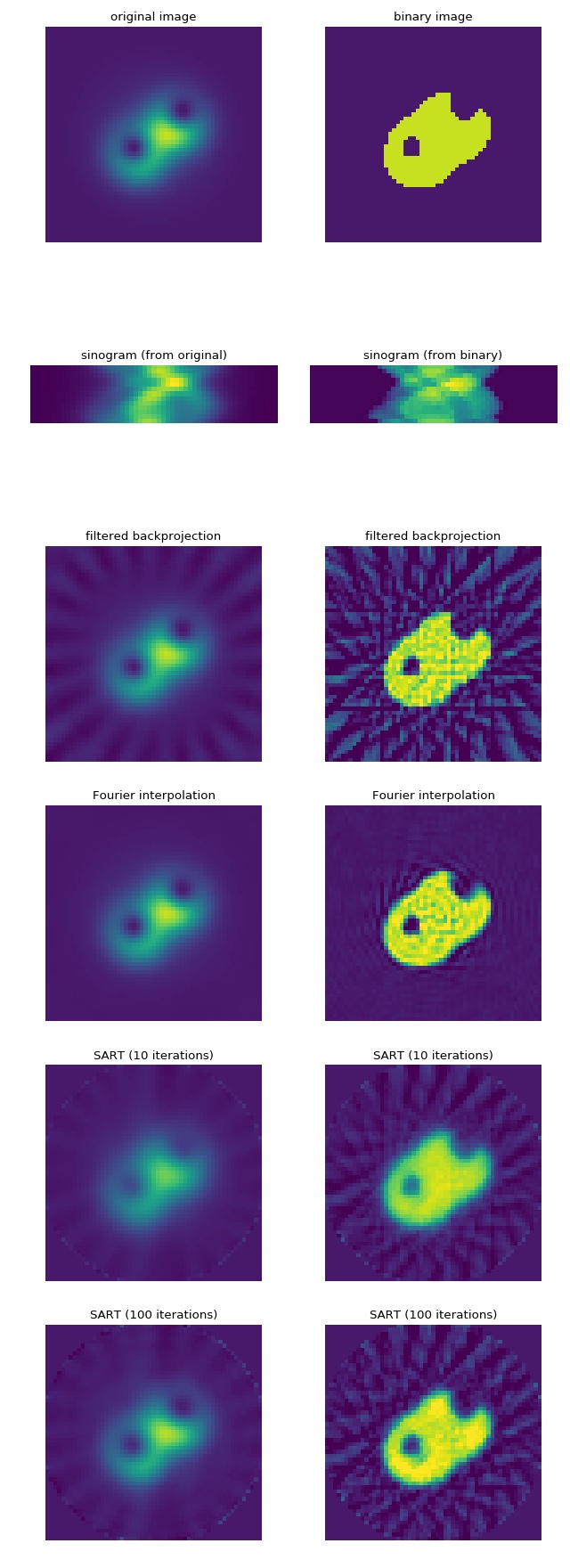
Comparison of parallel-beam reconstruction methods¶
This example illustrates the performance of the
different reconstruction techniques for a parallel-beam
geometry. The left column shows the reconstruction of
the original image and the right column shows the reconstruction
of the corresponding binary images. Note that the
SART process could be sped-up by computing an
initial guess with a non-iterative method and
setting it with the initial keyword argument.
1from matplotlib import pylab as plt
2import numpy as np
3
4import radontea
5from radontea.logo import get_original
6
7N = 55 # image size
8A = 13 # number of sinogram angles
9ITA = 10 # number of iterations a
10ITB = 100 # number of iterations b
11
12angles = np.linspace(0, np.pi, A)
13
14im = get_original(N)
15sino = radontea.radon_parallel(im, angles)
16fbp = radontea.backproject(sino, angles)
17fintp = radontea.fourier_map(sino, angles).real
18sarta = radontea.sart(sino, angles, iterations=ITA)
19sartb = radontea.sart(sino, angles, iterations=ITB)
20
21im2 = (im >= (im.max() / 5)) * 255
22sino2 = radontea.radon_parallel(im2, angles)
23fbp2 = radontea.backproject(sino2, angles)
24fintp2 = radontea.fourier_map(sino2, angles).real
25sarta2 = radontea.sart(sino2, angles, iterations=ITA)
26sartb2 = radontea.sart(sino2, angles, iterations=ITB)
27
28plt.figure(figsize=(8, 22))
29pltkw = {"vmin": -20,
30 "vmax": 280}
31
32plt.subplot(6, 2, 1, title="original image")
33plt.imshow(im, **pltkw)
34plt.axis('off')
35
36plt.subplot(6, 2, 2, title="binary image")
37plt.imshow(im2, **pltkw)
38plt.axis('off')
39
40plt.subplot(6, 2, 3, title="sinogram (from original)")
41plt.imshow(sino)
42plt.axis('off')
43
44plt.subplot(6, 2, 4, title="sinogram (from binary)")
45plt.imshow(sino2)
46plt.axis('off')
47
48plt.subplot(6, 2, 5, title="filtered backprojection")
49plt.imshow(fbp, **pltkw)
50plt.axis('off')
51
52plt.subplot(6, 2, 6, title="filtered backprojection")
53plt.imshow(fbp2, **pltkw)
54plt.axis('off')
55
56plt.subplot(6, 2, 7, title="Fourier interpolation")
57plt.imshow(fintp, **pltkw)
58plt.axis('off')
59
60plt.subplot(6, 2, 8, title="Fourier interpolation")
61plt.imshow(fintp2, **pltkw)
62plt.axis('off')
63
64plt.subplot(6, 2, 9, title="SART ({} iterations)".format(ITA))
65plt.imshow(sarta, **pltkw)
66plt.axis('off')
67
68plt.subplot(6, 2, 10, title="SART ({} iterations)".format(ITA))
69plt.imshow(sarta2, **pltkw)
70plt.axis('off')
71
72plt.subplot(6, 2, 11, title="SART ({} iterations)".format(ITB))
73plt.imshow(sartb, **pltkw)
74plt.axis('off')
75
76plt.subplot(6, 2, 12, title="SART ({} iterations)".format(ITB))
77plt.imshow(sartb2, **pltkw)
78plt.axis('off')
79
80
81plt.tight_layout()
82plt.show()
Volumetric data reconstruction benchmark¶
This simple example can be used to quantify the speed-up due to
multiprocessing on the hardware used. Note that
multiprocessing.cpu_count does not return the number of
physical cores.
1from multiprocessing import cpu_count
2import time
3
4import numpy as np
5
6import radontea as rt
7
8
9A = 70 # number of angles
10N = 128 # detector size x
11M = 24 # detector size y (number of slices)
12
13# generate random data
14sino0 = np.random.random((A, N)) # for 2d example
15sino = np.random.random((A, M, N)) # for 3d example
16sino[:, 0, :] = sino0
17angles = np.linspace(0, np.pi, A) # for both
18
19a = time.time()
20data1 = rt.backproject_3d(sino, angles, ncpus=1)
21print("time on 1 core: {:.2f} s".format(time.time() - a))
22
23a = time.time()
24data2 = rt.backproject_3d(sino, angles, ncpus=cpu_count())
25print("time on {} cores: {:.2f} s".format(cpu_count(), time.time() - a))
26
27assert np.all(data1 == data2), "2D and 3D results don't match"
Progress monitoring with progression¶
The progress of the reconstruction algorithms in radontea can be tracked with other packages such as progression.

1from multiprocessing import cpu_count
2
3import numpy as np
4import progression as pr
5
6import radontea as rt
7
8
9A = 55 # number of angles
10N = 128 # detector size x
11M = 24 # detector size y (number of slices)
12
13# generate random data
14sino0 = np.random.random((A, N))
15sino = np.random.random((A, M, N))
16sino[:, 0, :] = sino0
17angles = np.linspace(0, np.pi, A)
18
19count = pr.UnsignedIntValue()
20max_count = pr.UnsignedIntValue()
21
22
23with pr.ProgressBar(count=count,
24 max_count=max_count,
25 interval=0.3
26 ) as pb:
27 pb.start()
28 rt.volume_recon(func2d=rt.sart, sinogram=sino, angles=angles,
29 ncpus=cpu_count(), count=count, max_count=max_count)